National SARS-CoV-2 Whole Genome Sequencing (WGS) Surveillance Programme


Introduction
In 2021, an expert group was established to develop a proposal for a National SARS-CoV-2 Whole Genome Sequencing (WGS) Surveillance Programme in Ireland. The expert group’s proposal recommended that the Programme be developed and implemented, led by HSE-Health Protection Surveillance Centre (HPSC), with UCD National Virus Reference Laboratory (NVRL) as the lead partner laboratory. The primary aim of the Programme is to track the molecular epidemiology of SARS-CoV-2 in Ireland to inform and enhance the public health response to COVID-19. Associated programme goals include contributing to the global knowledge on SARS-CoV-2 to inform the global public health response and integrate SARS-CoV-2 genomics into routine public health practice.
How is the Programme structured?
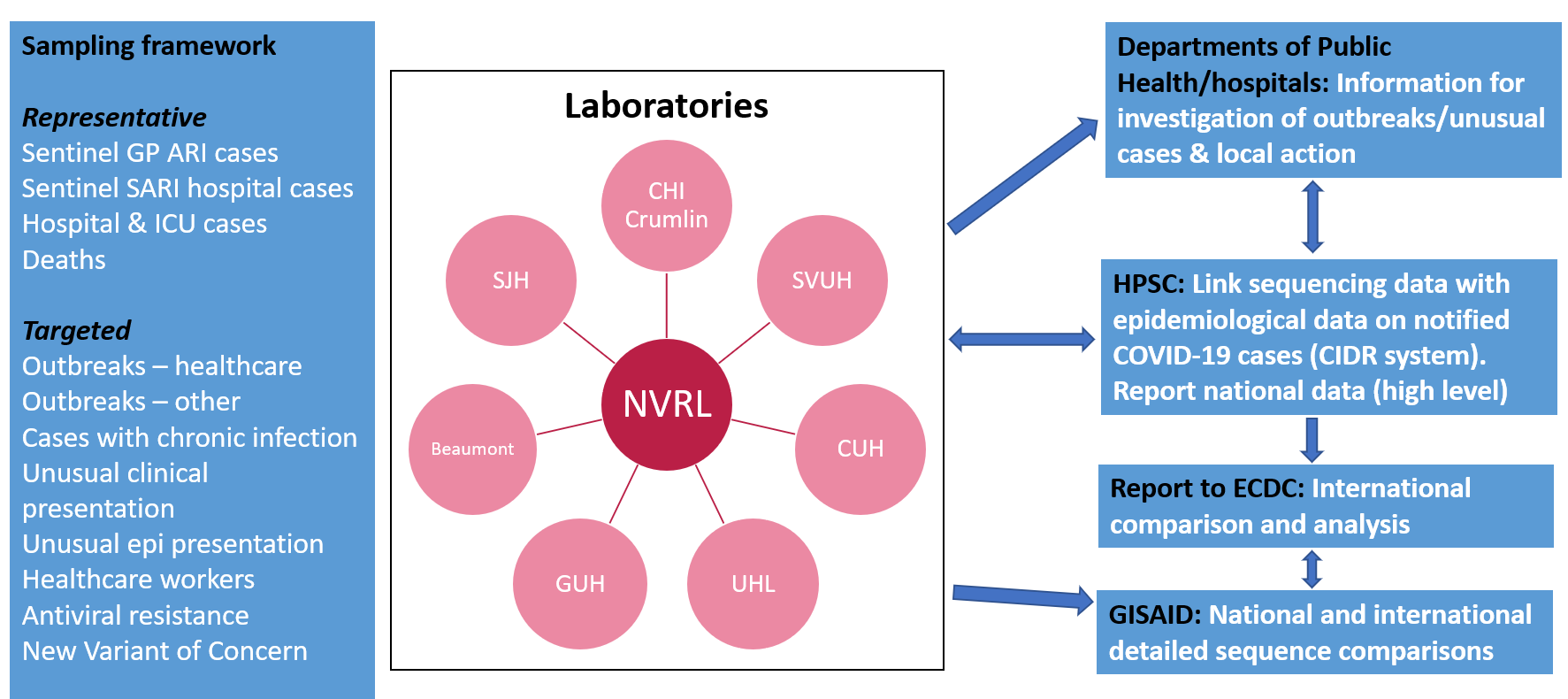
The Programme is led by HPSC with NVRL as the lead laboratory partner. The Programme Steering Group agreed a hub and spoke model for the Programme, incorporating NVRL (hub), six regional acute hospital laboratories (spokes), one national paediatric hospital spoke laboratory (Children’s Health Ireland-Crumlin) and HPSC as the surveillance partner. The following seven regional acute hospital laboratories are designated as spokes:
- University Hospital Limerick (UHL)
- St James’s Hospital (SJH)
- Galway University Hospitals (UHG)
- Beaumont Hospital
- Cork University Hospital (CUH)
- St Vincent’s University Hospital (SVUH)
- CHI-Crumlin paediatric hospital
These acute hospital spokes and CHI-Crumlin were selected to represent each of the six HSE Health Regions in line with public health reform and Sláintecare.
What are the Objectives of the National SARS-CoV-2 WGS Surveillance Programme?
The objectives of the National SARS-CoV-2 WGS Surveillance Programme, are:
- to collect WGS typing data i.e. Pango lineage on confirmed SARS-CoV-2 cases whose specimens have undergone WGS, and to record the data nationally
- to deposit SARS-CoV-2 genetic sequence data to the global SARS-CoV-2 sequence repository Global initiative on sharing all influenza Data (GISAID) platform in a timely manner
- to provide up to date information on the genomic epidemiology of SARS-CoV-2 for local and national incident and outbreak management
- to provide timely national reporting on the epidemiology of SARS-CoV-2 lineages, variants of concern or interest (and mutations of concern or interest) in Ireland to inform public health policy and actions
- to add to the global knowledge of SARS-CoV-2 epidemiology
How does the Programme work?
As the COVID-19 pandemic progressed and with the shift away from mass population testing to more targeted testing, SARS-CoV-2 genomic surveillance strategies have also become more focussed. Both the ECDC and WHO recommend that genomic sequencing for SARS-CoV-2 surveillance should target risk groups and individuals with moderate or severe symptoms as well as representative sentinel surveillance systems such as surveillance of acute respiratory infections (ARI) in primary care and severe acute respiratory infections (SARI) in hospital. They also stress the timely sharing of sequence data on public platforms such as GISAID for global monitoring of variant trends and early detection of emerging variants.
The Programme aims to sequence SARS-CoV-2 samples to monitor circulating variants, particularly those causing illness and severe disease, and to detect any variants with unusual characteristics and potential for increased transmissibility and outbreaks. The Programme implemented a Sampling Framework (see link below) that is based on sequencing cases from representative sentinel surveillance systems, and cases associated with severe disease and death, outbreaks in health and care settings, and changes in SARS-CoV-2 characteristics such as transmissibility and outbreak potential (Figure 1). It facilitates the timely reporting of SARS-CoV-2 WGS data to ECDC and/or other supra-national institutions, as well as sharing data in the public domain (e.g. in genetic sequence databases and repositories or epidemiological reports).
Figure 1: Overview of SARS-CoV-2 sequencing framework in Ireland.
Who governs the Programme?
The overall governance of the Programme is with the National SARS-CoV-2 WGS Programme Steering Committee.
How is the information shared?
The NVRL and spoke laboratories upload pseudonymised data on the SARS-CoV-2 sequences they generate to the GISAID platform. GISAID is the world’s largest repository for SARS-CoV-2 sequences.
Each laboratory uses bioinformatics software to process, quality check, and obtain detailed information on their sequences. These software analysis platforms use the Phylogenetic Assignment of Named Global Outbreak Lineages (PANGOLIN) software tool to compare the sequence to other sequences in its repository and to classify it into a SARS-CoV-2 lineage (Pango lineage). Laboratories then deposit the sequence data in GISAID whereby they are then provided with a GISAID generated accession number.
Sequence data uploaded on GISAID is accessible to registered GISAID users as well as international bodies such as the WHO and ECDC who perform regular analyses of sequences to monitor spatiotemporal trends and variant emergence. These analyses are shared in reports produced by these bodies as well as on publicly accessible platforms such as the ECDC/WHO European Respiratory Virus Surveillance Summary (ERVISS) website.
All participating laboratories report the assigned Pango lineage and GISAID Accession number to HPSC. The Pango lineage, GISAID ID and ‘Reason for sequencing’ information are linked and added to the case-based information notified on Ireland’s Computerised Infectious Disease Reporting (CIDR) system and the sequencing results can then be accessed by nominated CIDR users in HPSC and the local Departments of Public Health.
HPSC report nationally on the Pango lineage data in conjunction with epidemiological data on SARS-CoV-2 cases from the CIDR system. HPSC, the local Departments of Public Health and hospitals can also link with the NVRL and spoke laboratories in relation to more detailed analyses of the sequencing results where this is needed (e.g. specific variants of interest/concern or relatedness of cases in a suspected outbreak).
HPSC is also responsible for uploading COVID-19 data to the European Centre for Disease Prevention and Control (ECDC) online surveillance system - The European Surveillance System (TESSy). The pseudonymised data uploaded to TESSy include Pango lineage and GISAID ID in addition to epidemiological data. Access to TESSy is controlled and patient names, addresses and dates of birth are not included in the files uploaded.
HPSC produces publicly available surveillance reports of SARS-CoV-2 variant trends in Ireland. Variant trend analysis in Ireland is also shared with the following individuals and groups through reports and/or regular meetings: Director of HPSC, Director of National Health Protection, National Director of Public Health, HSE Chief Clinical Officer and other HSE leads, the Chief Medical Officer, Public Health and laboratory colleagues, Health Protection Respiratory Special Interest group and HPSC staff via numerous meetings including weekly scientific meetings, Health Protection meetings and Unscheduled Emergency Care meetings.
More information
You can contact the WGS Programme at WGS.programme@hpsc.ie
Last Updated: 10 May 2024


